SSomerville

- Principal Investigator
PhD Agronomy, Plant Physiology University of Illinois, Urbana-Champaign, 1981
B.S. Genetics University of Alberta, 1976
We study plant-pathogen interactions, especially the host's active, if unwitting, role in disease development. We work with powdery mildew disease on model plant Arabidopsis thaliana, using mutational analysis to identify host factors required for successful disease development. We also study a new area of plant-pathogen biology, non-host resistance that protects all members of a plant species from all members of a pathogen species. Results from both these projects highlight the importance of both active and passive defenses operating in the host cell wall.
Plant-pathogen interactions
Traditionally, research in the field of plant pathology has focused on incompatible interactions - deciphering the host components associated with disease resistance and the pathogen factors that trigger host defenses. More recently, increasing attention has been focused on host and pathogen factors that promote compatible interactions and disease development (Chrisholm et al. 2006).

Powdery mildew infection, cp = conidiophores
We study the interactions between the model host plant Arabidopsis thaliana and the obligate fungal pathogen Golovinomyces cichoracearum, the causal agent of the powdery mildew disease on Arabidopsis. Because the powdery mildew pathogens have a biotrophic lifestyle, requiring living plant tissues for survival, we reasoned that the host plant is likely to be an active partner in compatible interactions leading to disease. Ways in which the host might contribute to disease development include the formation of structural components of the haustorial complex, such as the extrahaustorial membrane. Also, since the powdery mildew pathogen draws water, mineral and organic nutrients from its host via the haustorium, adjustments in host metabolism to accommodate the additional demand by the pathogen are likely to be required.
We have taken several approaches to determining host contributions to the powdery mildew disease development. Descriptive experiments to define changes in transcript levels using DNA microarrays and changes in subcellular compartments as monitored in Arabidopsis lines carrying GFP-marked organelles have been conducted. In addition, mutational strategies to identify host susceptibility genes by their ability to limit powdery mildew growth when mutated have been conducted.
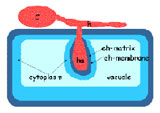
Cartoon of powdery mildew-infected epidermal cell with haustorium (ha), a fungal feeding structure, in an extrahaustorial matrix (eh) encased in the extrahaustorial membrane conidium, cn.
Reference:
Chisholm, S.T., G. Coaker, B. Day and B.J. Staskawicz. 2006 Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803-14.
Powdery Mildew Resistant (pmr) Mutants
To identify host susceptibility factors, a direct screen for loss-of-susceptibility mutants was conducted. Such mutants appear powdery mildew resistant and can be readily identified among susceptible (wild-type) individuals 7-9 days post-inoculation with the powdery mildew pathogen. The mutants from the screen (n=26) were placed into six complementation groups. In each case, the mutant gene was genetically recessive suggesting that the mutation could be "loss-of-function" mutation. All six of the mutant classes have been described (Nishimura et al. 2003, Vogel and Somerville 2000, Vogel et al. 2002, Vogel et al. 2004).
In a collaborative project with Ralph Panstruga (Max Planck Institute, Cologne, Germany), we discovered that PMR2 (=MLO2) is the primary functional orthologue of the barley gene MLO, a negative regulator of defenses against the barley powdery mildew disease (Consonni et al. 2006). Two related Arabidopsis genes, MLO6 and MLO12 appear to play a minor supporting role to MLO2. Loss-of-function mutants of the barley MLO gene are powdery mildew resistant and Arabidopsis mlo2 mutants exhibit many of the same features as their barley counterparts. The defenses elicited in mlo mutants act early in the infection sequence primarily to limit penetration by the powdery mildews into epidermal cells. Interestingly, mutations in the PEN1 (syntaxin), PEN2 (glycosyl hydrolase) or PEN3 (ABC transporter) suppress penetration resistance in mlo2 mutants. As noted under the section on Non-Host Resistance, the pen1-pen3 Arabidopsis mutants were recovered in a screen for mutants compromised in resistance to inappropriate pathogens (e.g., Arabidopsis resistance to Blumeria graminis f. sp. hordei, barley powdery mildew). Thus, it would appear that the resistance mechanisms under negative control by MLO contribute to resistance to both inappropriate and appropriate pathogens.
The PMR4 gene was found to encode a wound- and pathogen-associated callose synthase (Nishimura et al. 2003). Callose deposition in cell wall appositions, called papillae, just below penetration sites is a common defense response in plants. Thus, it was unexpected that plants deficient for the wound- and stress-inducible callose synthase would be highly disease resistant. We discovered that the basis for this resistance was a hyper-activation of the salicylic acid defense pathway in infected pmr4 plants. How a deficiency in the stress-inducible callose synthase and callose production de-represses (or activates) the salicylic acid pathway remains to be discovered.
The pmr5 and pmr6 mutants are very similar morphologically. The PMR6 gene encodes a novel pectate lyase-like protein and is predicted to localize to the outer surface of the plasma membrane (Vogel et al. 2002), while the PMR5 gene encodes a novel protein predicted to be in the endoplasmic reticulum (Vogel et al. 2004). Double mutant analysis demonstrated that pmr5- and pmr6-based resistances operate independently of the known defense signal transduction pathways (i.e., the salicylic acid and the ethylene/jasmonate pathways). The predicted function of PMR6 as a cell wall degrading enzyme led to an analysis of the cell walls in pmr5 and pmr6. Fourier transform infrared spectrometry indicated that the pectin composition of cell walls from both mutants was altered, suggesting that these two genes participate in pectin biosynthesis, modification or deposition in some manner. Pectin components play important roles in determining the porosity and ionic environment of the cell wall, both features that could influence the exchange of signaling molecules and nutrients between host and pathogen (Vincken et al. 2003). Furthermore, some pectin polymers contain latent elicitors that can be released by pathogen cell wall degrading enzymes during the penetration step (Vorwerk et al. 2004). Thus, the loss of PMR5 or PMR6 may lead to the activation of a novel defense pathway. Alternatively, the powdery mildew pathogen may not longer properly recognize the pmr5 or pmr6 plants as suitable hosts or the powdery mildew may not be able to efficiently penetrate the epidermal wall due to the change in cell wall. An active area of interest in the lab is to determine which of these hypothesized mechanisms might confer resistance in the pmr5 and pmr6 mutants.

Phenotypes of pmr mutants. L to R: pmr1, pmr2, pmr3, pmr4, pmr5, and pmr6, shown 10 days after inoculation with the Arabidopsis powdery mildew, G. cichoracearum.
References:
Consonni, C., M.E. Humphry, H.A. Hartmann, M. Livaja, J. Durner, L. Westphal, J. Vogel, V. Lipka, B. Kemmerling, P. Schulze-Lefert, S.C. Somerville and R. Panstruga. 2006. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nature Genetics 38, 716-720.
Nishimura, M., M. Stein, B-H. Hou, J. Vogel, H. Edwards and S. Somerville. 2003. Loss of a callose synthase results in salicylic acid-dependent disease resisitance. Science 301: 969-972
Vincken, J.P., H.A. Schols, R.J.F.J. Oomen, M.C. McCann, P. Ulvskov, A.G.J. Voragen and R.G.F. Visser. 2003. If homogalacturonan were a side chain of Rhamnogalacturonon I. Implications for cell wall architecture. Plant Physiology 132, 1781-1789.
Vogel, J. and S.C. Somerville. 2000. Isolation and characterization of powdery mildew resistance Arabidopsis mutants. Proceedings of the National Academy of Sciences, USA 97: 1897-1902.
Vogel, J.P., T.K. Raab, C. Schiff and S. Somerville. 2002. PMR6, a pectate-lyase like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095-2106.
Vorwerk, S., S. Somerville and C. Somerville. 2004. The role of plant cell wall polysaccharide composition in disease resistance. Trends in Plant Science 9, 203-209.
Powdery Mildew-Induced Lesion (mil) Mutants

A variety of abiotic stresses, such as wounding, heat and cold stress, or drought, failed to elicit necrosis or chlorosis in mil1 or mil4 mutants. Inoculation with the virulent bacterial pathogen, Pseudomonas syringae pv. tomato DC3000 did not elicit lesion formation. Among the various treatments, only inoculation with the inappropriate pathogen, barley powdery mildew, also induced necrosis several days after inoculation. The MIL1 (=EDR2) gene was cloned by T-DNA tagging and found to encode a novel gene with three predicted distinct domains -- a StAR (steroidogenic acute response protein) transfer domain, which is a lipid/sterol-binding domain; a pleckstrin homology domain binds to phosphatidylinositol-4-phosphate; and a plant-specific domain of unknown function (DUF1336) (Tang et al. 2005). The MIL4 gene encodes a novel small protein with four transmembrane domains and two splice variants.
It is notable that many of the classic components of programmed cell death in animal systems cannot be readily identified among plant genes. Thus, it would appear that plants may have evolved independent mechanisms for regulating and executing cell death during development and in response to environmental factors. The MIL genes may help us understand the cell death program triggered during plant-pathogen interactions.
Reference:
Tang, D., J. Ade, C.A. Frye and R.W. Innes. 2005. Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. The Plant Journal 44, 245-257.
Subcellular Responses to Powdery Mildew Infection
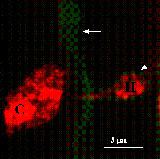
A few previous studies with epidermal peels monitored changes in subcellular components in living plant tissue but these observations were limited to those that could be made with Nomarski optics in the absence of staining (see review Aist and Bushnell 1991). In agreement with this literature, GFP-tagged organelles accumulated around the haustorial complex (Koh et al. 2005). This was most dramatic with the peroxisomes, which appear to move preferentially toward haustoria. However, all other subcellular structures appeared to aggregate near the haustorium. In addition, the GFP-plasma membrane marker lines dramatically illustrate that the extrahaustorial membrane, the plant membrane encasing the haustorial complex, is distinct from the plasma membrane (Koh et al. 2005). How the extrahaustorial membrane is formed and what the permeability properties of this membrane are will be important questions to address in future experiments, as the formation of this specialized membrane is key to successful fungal infection.
References:
Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and C.R. Somerville. 2000. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proceedings of the National Academy of Sciences, U.S.A. 97: 3718-3723.
Koh, S., A. André, H. Edwards, D. Ehrhardt and S. Somerville. 2005. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. The Plant Journal 44, 516-529.
NON-HOST RESISTANCE IN ARABIDOPSIS
Non-host resistance is an operational definition that describes the resistance observed when all members of a plant species exhibit resistance to all members of a given pathogen species (Heath 1991, Thordal-Christensen 2003). This form of resistance is rarely studied by comparison with the more familiar race-cultivar specific resistance employed by plant breeders in developing disease resistant cultivars. A number of race-cultivar specific resistance genes have been cloned and found to encode proteins that have the hallmarks of receptors, features which are consistent with long-standing models of the function of host resistance proteins. In contrast, non-host resistance is thought to comprise a variety of distinct mechanisms that may include the production of pre-formed toxins or barriers, or a lack of essential metabolites or signaling molecules required by the pathogen. Thus, non-host resistance is thought to be multi-genic and the inactivation of any one component may not be sufficient to render a plant susceptible. A further component of the model developed by M. Heath (1991) is that basic compatibility is established in an incremental process over evolutionary time by the subversion of non-host defense mechanisms by a pathogen species.
Our group has characterized the non-host responses of Arabidopsis to the barley powdery mildew pathogen, Blumeria graminis f. sp. hordei. A majority of fungal propagules were arrested at the penetration stage and never formed haustoria (Yun et al. 2003, Zimmerli et al. 2004). However, a small percentage (~5%) were able to form haustoria and in some cases elongating secondary hyphae (a measure that the haustoria are functional). No conidiophores or asexual conidia developed. After surveying a variety of host responses to inoculation with B. g. hordei (e.g., papilla formation, callose deposition, H2O2 staining, dead cells), the most dramatic difference between responses to the barley and Arabidopsis powdery mildews was the callose response. Both fungi elicited papilla formation at sites of attempted penetration; however, the barley powdery mildew also elicited wide-spread deposition of callose in mesophyll cells subtending infected epidermal cells (Zimmerli et al. 2004).
A screen for Arabidopsis mutants with an altered callose response to B. g. hordei was initiated. The screen was designed to recover both callose-deficient and callose-enhanced mutants. To date, mutants in four pen (penetration resistance) complementation groups have been recovered. Two of the mutant classes, pen1 and pen2, were previously identified in other screens for defects in non-host resistance (Collins et al. 2003; Lipka et al. 2005). PEN1 encodes a syntaxin (SYP122) and PEN2 a glycosyl hydrolase. PEN3 (=PDR8) encodes an ABC transporter of the pleiotropic drug resistance class (Stein et al. 2006). Although the specific roles of each of these components in penetration resistance is unknown, our working hypothesis is that PEN2 participates in the synthesis of a toxic secondary metabolite and that PEN1 and PEN3 export materials to the cell wall space to block pathogen ingress. These defenses substances may be toxic compounds or materials to reinforce the cell wall (Stein et al. 2006).
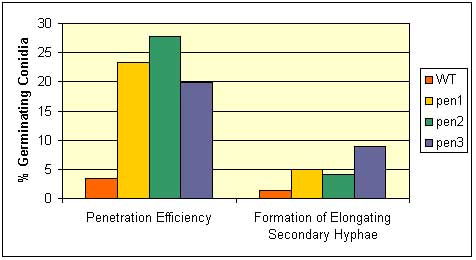
Fig. 1. Penetration and formation of elongating secondary hyphae by the barley powdery mildew pathogen on three pen mutants. Left panel (1): % of germinated conidia that penetrate the host cell wall, Right panel (2): % of germinated conidia that form elongating secondary hyphae, a measure of successful establishment of haustoria (fungal feeding structure). Red, wild-type; Yellow, pen1 (48M3); green, pen2 (136N4); blue, pen3 (114N4). From Stein et al. 2006.
References:
Collins, N., H. Thordal-Christensen, V. Lipka, S. Bau, E. Kombrink, M. Stein, R. Hückelhoven, S. Somerville and P. Schulze-Lefert. 2003. Conserved SNARE secretion machinery components mediate cell wall penetration resistance against powdery mildew plant pathogens. Nature 425, 973-977.
Heath, M.C. 1991. The role of gene-for-gene interactions in the determination of host species specificity. Phytopathology 81: 127-130.
Lipka, V., J. Dittgen, P. Bednarek, R. Bhat, M. Wiemer, M. Stein, J. Landtag, W. Brandt, S. Rosahl, D. Scheel, F. Llorente, A. Molina, J. Parker, S. Somerville and P. Schulze-Lefert. 2005. Pre- and post-invasion defenses both contribute to nonhost resistance in Arabidopsis. Science 301, 1180-1183.
Stein, M., J. Dittgen, C. Sánchez-Rodriguez, B.-H. Hou, A. Molina, P. Schulze-Lefert, V. Lipka and S. Somerville. 2006. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. The Plant Cell 18, 731-746.
Thordal-Christensen, H. 2003. Fresh insights into processes of nonhost resistance. Current Opinion in Plant Biology 6: 351-357.
Yun, B.W., H.A. Atkinson, C. Gaborit, A. Greenland, N.D. Read, J.A. Pallas and G.J. Loake. 2003. Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant Journal 34: 768-777.
Zimmerli, L., M. Stein, V. Lipka, P. Schulze-Lefert and S. Somerville. 2004. Host and nonhost pathogens elicit different jasmonate/ethylene response in Arabidopsis. The Plant Journal 40, 633-646.
Gene Expression Profiling of Nonhost Resistance
A comparison of the expression profiles of Arabidopsis plants infected with the barley powdery mildew (incompatible, non-host interaction) to those infected with the Arabidopsis powdery mildew, Golovinomyces cichoracearum (compatible interaction) showed that a majority of transcripts were either induced or repressed by both types of pathogens, even though the outcomes of the infections were markedly different (Zimmerli et al. 2004). Among the transcripts that were repressed by infection with either pathogen are a number of photosynthesis and carbon metabolism-related transcripts. This repression is thought to represent a reallocation of resources to defense responses. A number of common defense-related transcripts were up-regulated in response to infections by either pathogen and may represent defense genes associated with basal resistance, a form of resistance with limited effectiveness against the Arabidopsis powdery mildew. A relatively small number of genes were differentially expressed or expressed more rapidly following infections with B. graminis f.sp. hordei.
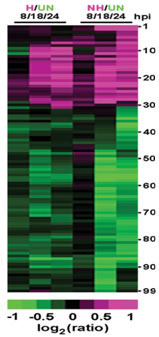
Fig. 2. Comparison of the gene expression profiles of Arabidopsis plants inoculated with a host pathogen or a nonhost pathogen. Three time points were monitored: 8, 18 and 24 hours post-inoculation. H: plants inoculated with the Arabidopsis powdery mildew, G. cichoracearum; NH: plants inoculated with the barley powdery mildew, B. g. hordei; UN: uninoculated control plants. From Zimmerli et al. 2004.
Reference
Zimmerli, L., M. Stein, V. Lipka, P. Schulze-Lefert and S. Somerville. 2004. Host and nonhost pathogens elicit different jasmonate/ethylene responses in Arabidopsis. (in revision).
Chito-oligomer-Induced Changes in Gene Expression Interaction
Chitin is a common component of fungal and insect cell walls and is likely to be one of the signals exchanged between host and pathogen that impact the outcome of an infection. Chito-oligomers are known to elicit defense responses, such as the production of reactive oxygen species and the induction of defense gene expression. Because of the ubiquity of chitin in pathogenic and non-pathogenic fungi and insects, chito-oligomers are likely to be non-specific elicitors that may play a role in triggering non-host defenses or basal defenses. In collaboration with Gary Stacey (University of Missouri, Columbia, MO), we have generated a profile of the Arabidopsis genes induced or repressed by treatment with chito-oligomers (Ramonell et al. 2002, Ramonell et al. 2004, Zhang et al. 2002). A number of transcripts were up-regulated at the earliest time point, 10 min, with the largest number of changes in transcript levels occurring at 30 min after chito-oligomer treatment. Comparisons with the transcript profiles of salicyclic acid-, ethylene- or jasmonate-treated plants suggested that chito-oligomers do not act via these well-known signaling pathways.
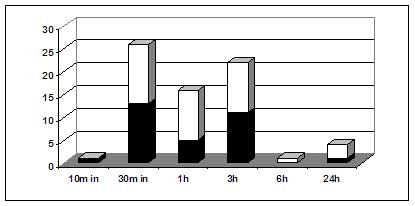
Fig. 3. Number of differentially expressed genes at various times following chito-oligomer treatment. Dark boxes, down-regulated genes. White boxes, up-regulated genes. From: Ramonell et al. 2002. Insertion mutants of three chito-oligomer responsive genes, two disease resistance gene-like genes and an E3 ligase gene, were shown to be mildly compromised in their resistance to the Arabidopsis powdery mildew pathogen (Ramonell et al. 2004). These results confirm that chito-oligomer-based signaling does contribute to resistance to powdery mildew.
References:
Ramonell, K.M., B. Zhang, R.M. Ewing, Y. Chen, D.Xu, G. Stacey and S. Somerville. 2002. Microarray analysis of chitin elicitation in Arabidopsis thaliana. Molecular Plant Pathology 3: 301-311.
Ramonell, K., M. Berrocal-Lobo, S. Koh, J. Wan, H. Edwards, G. Stacey and S. Somerville. 2005. Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen, Erysiphe cichoracearum. Plant Physiology 138, 1027-1036.
Zhang, B., K. Ramonell, S. Somerville and G. Stacey. 2002. Characterization of early, chitin-induced gene expression in Arabidopsis. Molecular Plant-Microbe Interactions 15: 963-970.
Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, Somerville S, Wu SH.LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation.Plant J. 2008 Apr;54(2):205-19. Epub 2008 Jan 7.
Zimmerli L, Hou BH, Tsai CH, Jakab G, Mauch-Mani B, Somerville S.The xenobiotic beta-aminobutyric acid enhances Arabidopsis thermotolerance.Plant J. 2008 Jan;53(1):144-56. Epub 2007 Nov 28.
Espinoza C, Medina C, Somerville S, Arce-Johnson P.Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis.J Exp Bot. 2007;58(12):3197-212. Epub 2007 Aug 30. article
Vorwerk S, Schiff C, Santamaria M, Koh S, Nishimura M, Vogel J, Somerville C, Somerville S.EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana.BMC Plant Biol. 2007 Jul 6;7:35. article
Hernández-Blanco C, Feng DX, Hu J, Sánchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sánchez-Rodríguez C, Anderson LK, Somerville S, Marco Y, Molina A.Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance.Plant Cell. 2007 Mar;19(3):890-903. Epub 2007 Mar 9. article
Schenk PM, Kazan K, Manners JM, Anderson JP, Simpson RS, Wilson IW, Somerville SC, Maclean DJ "Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola." Plant Physiol 2003; 132: 2: 999-1010
Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, Schulze-Lefert P "SNARE-protein-mediated disease resistance at the plant cell wall." Nature 2003; 425: 6961: 973-7
Cartieaux F, Thibaud MC, Zimmerli L, Lessard P, Sarrobert C, David P, Gerbaud A, Robaglia C, Somerville S, Nussaume L "Transcriptome analysis of Arabidopsis colonized by a plant-growth promoting rhizobacterium reveals a general effect on disease resistance." Plant J 2003; 36: 2: 177-88
Scheible WR, Fry B, Kochevenko A, Schindelasch D, Zimmerli L, Somerville S, Loria R, Somerville CR "An Arabidopsis mutant resistant to thaxtomin A, a cellulose synthesis inhibitor from Streptomyces species." Plant Cell 2003; 15: 8: 1781-94
Vogel JP, Raab TK, Schiff C, Somerville SC "PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis." Plant Cell 2002; 14: 9: 2095-106
Finkelstein D, Ewing R, Gollub J, Sterky F, Cherry JM, Somerville S "Microarray data quality analysis: lessons from the AFGC project. Arabidopsis Functional Genomics Consortium." Plant Mol Biol 2002; 48: 1-2: 119-31
Nishimura M, Somerville S "Plant biology. Resisting attack." Science 2002; 295: 5562: 2032-3
Zhang B, Ramonell K, Somerville S, Stacey G "Characterization of early, chitin-induced gene expression in Arabidopsis." Mol Plant Microbe Interact 2002; 15: 9: 963-70
Ramonell, K.M., B. Zhang, R.M. Ewing, Y. Chen, D. Xu, G. Stacey, S. Somerville "Microarray analysis of chitin elicitation in Arabidopsis thaliana" Molecular Plant Pathology 2002; 3: 301-311
Stein M, Somerville SC "MLO, a novel modulator of plant defenses and cell death, binds calmodulin." Trends Plant Sci 2002; 7: 9: 379-80
Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES "Expression profile analysis of the low-oxygen response in Arabidopsis root cultures." Plant Cell 2002; 14: 10: 2481-94
Ramonell KM, Somerville S "The genomics parade of defense responses: to infinity and beyond." Curr Opin Plant Biol 2002; 5: 4: 291-4
Wilson IW, Schiff CL, Hughes DE, Somerville SC "Quantitative trait loci analysis of powdery mildew disease resistance in the Arabidopsis thaliana accession kashmir-1." Genetics 2001; 158: 3: 1301-9
Schiff, C.L., I.W. Wilson, S.C. Somerville "Polygenic powdery mildew disease resistance in Arabidopsis thaliana. Quantitative trait analysis of the accession Warschau-1" Plant Pathology 2001; 50: 690-701
Zhou F, Kurth J, Wei F, Elliott C, Valè G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P "Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway." Plant Cell 2001; 13: 2: 337-50
Wu, S.-H., K. Ramonell, J. Gollub, S. Somerville "Plant gene expression profiling with DNA microarrays" Plant Physiology and Biochemistry 2001; 39: 917-926
Richmond T, Somerville S "Chasing the dream: plant EST microarrays." Curr Opin Plant Biol 2000; 3: 2: 108-16
Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM "Coordinated plant defense responses in Arabidopsis revealed by microarray analysis." Proc Natl Acad Sci U S A 2000; 97: 21: 11655-60
Vogel J, Somerville S "Isolation and characterization of powdery mildew-resistant Arabidopsis mutants." Proc Natl Acad Sci U S A 2000; 97: 4: 1897-902
170 - Modern Applications of Plant Biotechnology
299 - Graduate Research
